Cryogenic Electron Microscopy Method Assists in COVID-19 Vaccine Development
Members can download this article in PDF format.
What you’ll learn:
- How has cryogenic electron microscopy (cryo-EM) aided researchers during the COVID-19 pandemic?
- Applications of Camera Link HS (CLHS) outside the factory floor.
Advances in imaging technology continue to benefit society in disparate ways. For example, cryogenic electron microscopy (cryo-EM) played a fundamental role in the development of the COVID-19 vaccine. Combining cameras and detectors, sample-handling technology, automation, and software, cryo-EM is an easy-to-use method for high-quality data collection. It ultimately helped researchers identify the spike protein of SARS-CoV-2, the virus that causes COVID-19.
A Long, Automated Process
Cryo-EM (Fig. 1) involves flash-freezing a sample down to –180°C, leaving the protein particles suspended in ice. Sample sets are placed in a holder containing a grid that’s typically made of gold or carbon. Within the grid is foil with small holes or pockets. Material that’s imaged by the system is flash-frozen inside fluid contained in these holes.
To start the process, researchers use EPU 2 software from Thermo Fisher Scientific to define acquisition and optics settings as well as verify proper microscope alignment for high-resolution data acquisition. The software then acquires a grid overview to determine whether grids are of sufficient quality for an automated recording session. The grids are batch screened, with the software automatically grouping grid squares into categories of similar quality.
Lastly, researchers select grid squares and foil holes for acquisition and define the acquisition template for each foil hole. Grid-square and foil-hole selection can be done either manually or automatically by applying user-defined filter settings. The system uses an electron beam to illuminate individual holes, allowing the camera to capture an image. From there, the cryo-EM system moves the beam to multiple locations in the hole or to multiple holes and then the stage moves, enabling the beam to move to the next location. The beam effectively moves across multiple locations at each sample hole.
Because the process uses low electron doses to avoid damaging samples, and to allow for the detection of individual electron impacts, the images come out noisy and have low contrast. The system must acquire thousands of frames, which are aligned and summed to produce a suitable image. The reconstruction software also removes noise.
“Inspecting individual frames would not allow a system operator to make anything out in the image because of the low level of detail, so you’d just see noise,” said Hans Roeven, staff engineer for systems design at Thermo Fisher Scientific.
“The system might combine one or two thousand individual frames to produce an image with sufficient contrast that can be fed into the reconstruction process,” he continues. “Creating an entire 3D reconstruction requires tens of thousands of such images, which is why the process requires so much time. Increased camera speed (frame rate) plays an important role in reducing the overall data-collection time, thereby easing the workflow and optimizing time to results for our customers.”
Cryo-EM and COVID-19
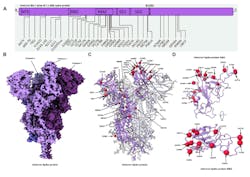
Single-particle cryo-EM involves taking particles extracted from thousands of acquired images to construct 3D representations of viruses or proteins (Fig. 2). This process produced the first 3D structures of the isolated spike protein of SARS-CoV-2. The spike protein is a key point of engagement with other cells. It binds with human host cell receptors before beginning the process of virus replication.
Disabling the spike protein’s binding ability is crucial according to Eric Chen, director of market development for Asia and the Pacific at Thermo Fisher Scientific. Prior to its identification, the spike protein was a focus for vaccine development. To understand its structure and produce an epitope map of binding sites, researchers sought to image it. This would allow them to determine which antibodies or inhibitors would prevent viral docking, said Chen.
To learn more about the spike protein, a team from the University of California-Berkeley recently used Thermo Fisher Scientific’s Krios G4 cryo-EM instrument equipped with a cold field emission gun (CFEG) operating at 300 kV. Used for image capture was a Falcon 4 detector, which features a custom-designed 4096 × 4096 CMOS image sensor and an internal frame rate of 250 fps (Fig. 3). It was back-thinned to 30 µm to ensure that the electron hits didn’t generate too large a cloud of electrons, which would appear as a large block in an image. Electron impact and captured images were read out with an FPGA through a Camera Link HS (CLHS) interface; its data-acquisition system was comprised of custom FPGA boards.
Leveraging cryo-EM technology, UC Berkeley researchers found that the spike protein is nothing like other known ion channels: It consists of half a channel, piercing the cell membrane only halfway. The SARS-CoV-2 ORF3a protein is a putative viral ion channel implicated in autophagy inhibition, inflammasome activation, and apoptosis. The 3a protein and anti-3a antibodies are found in infected tissues and plasma, according to UC Berkeley team’s study, which was published in Nature.
Researchers have found that deletion of 3a in SARS-CoV-2 and deletion of a related 3a gene in the original SARS virus, SARS-CoV-1, reduces the severity of illness in animal models. The UC Berkeley researchers noted that targeting this protein in the vaccine helped reduce the severity of human COVID-19 infections.
CLHS Tackles Data Challenges
While most often associated with machine-vision applications, the Association for Advancing Automation’s (A3) CLHS standard offers benefits that extend far beyond the factory floor. Thermo Fisher Scientific leverages the CLHS standard in its transmission-electronic-microscopy (TEM) cameras and detectors, including the system used to identify the spike protein.
TEM applications involve high-resolution, high-speed cameras that generate massive amounts of data (images). While Thermo Fisher Scientific has used cameras and detectors for a long time, reducing overall data-collection time represents a significant factor in new product development, according to Roeven.
“Workflow automation and image acquisition speed are important factors, and new camera developments involve ever-increasing data rates, so robust, high-speed interfaces are critical to success,” he said. “We not only wanted to standardize the interface, but also required low-latency triggering to synchronize processes in the microscope, which CLHS delivers.”
Clocking resources into the FPGA presented some challenges during development, since the company combines up to eight lanes in a single FPGA. Specifically, the interface needed to fit in a design that uses several resources for other functions. This required the careful partitioning of clock resources for adding an eight-lane, 10-Gb/s interface.
In turn, the system can effectively share resources for multiple instances of the CLHS X-Protocol IP core—64b/66b line encoding with forward error correction designed for 10-Gb/s and higher bit rates. “In addition to the crucial task of synchronization, the low-latency, zero-jitter link trigger offered by CLHS gave us more freedom in partitioning functions and allowing the system to run at full potential,” said Roeven.
Using fiber cables in the system provides galvanic isolation to avoid ground faults in the system, while also offering cost-effectiveness, since FPGAs with QSFP+ interfaces are generally available off-the-shelf. In addition, according to Roeven, fiber cables help the system avoid image degradation via noise.
“Using fiber also allows us to place the image-processing system at an acceptable distance from the instrument, which is important because computer servers can make a lot of noise, and acoustic vibrations can be a source of image degradation when resolving images down to such levels,” said Roeven.
Future Innovations
While cryo-EM methods have gained a lot of attention for their use in COVID-19 vaccine development, the technology has already been popular for several years. Its creators won the Nobel Prize for Chemistry in 2017 for “developing cryo-electron microscopy for the high-resolution structure determination of biomolecules in solution.” The process was named Method of the Year 2015 by Nature.
Cryo-EM systems also are used in drug discovery, plant biology research, pathology research, cancer research, material sciences, and semiconductor inspection. Moving forward, the technology is something to keep an eye on, as it could have significant impacts in life sciences and beyond.